Abstract
As remarkable advances have been made in immunotherapies, the overall goal of immunotherapy has become the selection of patients and evaluating the benefits of treatment. One of the major obstacles to develop immunotherapies is the lack of effective immune monitoring. Monitoring of key changes in the immune system during immunotherapy (immunomonitoring) provides important insights into efficacy as well as the immune mechanisms of response at the molecular and cellular levels. Immunomonitoring techniques include traditional immunoassays that use specific antibodies to recognize the analytes of interest, new high-throughput immunoassays that target immune cells and nucleic acids, and less classical immunogenomic approaches that rely on genome-wide profiling and computational analysis on various types of clinical samples. Substantial progress has been made in the application of immunomonitoring strategies to pre-clinical and clinical studies, especially for patients with cancer and infectious diseases. Current and emerging immunoassays performed in clinical practice will be examined herein, and immunogenomic approaches that complement these techniques will be highlighted and compared with traditional methods. Finally, we will discuss several new computational methods for analyzing gene signatures for immunomonitoring, including gene expression data profiling by microarray, the nCounter technique, regular RNA-seq, and single-cell RNA-seq. Novel immunomonitoring techniques, especially immunogenomic approaches, will continue to be developed to facilitate assessment of immunotherapeutic response and predict patient outcomes in cancer and infectious disease.
Author Contributions
Academic Editor: Hou YC, The Lab of Tumor Molecular Cellular Biology, Shaanxi Normal University Xian, Shaanxi, China.
Checked for plagiarism: Yes
Review by: Single-blind
Copyright © 2019 Xi Zhang, et al.
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Introduction
As remarkable clinical success have been made in immunotherapies, immunotherapy has been established as a powerful treatment option in cancer. Newly emerging immunotherapies, including antibodies against cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), programmed cell death protein 1 (PD-1), and programmed death ligand 1 (PD-L1), have demonstrated notable efficacy in several types of cancer 1. Currently only a small group of patients benefit from these therapies, and the mechanisms of primary and acquired resistance to these therapies remain poorly understood. A better understanding is needed of the cellular and molecular factors that modulate the sustained immune response after the initial response to immunotherapy 2.
Immune monitoring (immunomonitoring) of key changes in the immune system can provide important insights into the mechanisms that determine therapeutic response at the molecular and cellular levels 3. Immune-related adverse events, cytokine and immune cell responses, and overall survival benefit outcomes of interest in immunomonitoring studies. Highly specialized immunoassays have evolved in the past two decades, and have been applied to immunomonitoring in human clinical trials 4, 5. These techniques are useful not only in dissecting the dynamic changes in the tumor microenvironment, but also in evaluating immune cell composition in patients with infectious and other diseases and the effect of therapy on the immune response 6, 7. Thus far, changes in the composition of immune cell subsets has been investigated with classical immunomonitoring techniques, including immunofluorescence, flow cytometry, and mass cytometry. Recent developments in next-generation sequencing (NGS) technologies has led to applications in routine immunomonitoring 8. For example, the composition of tumor-infiltrating immune cells can be characterized from bulk tumor RNA-seq data using computational approaches based on a set of immune-specific marker genes or expression signatures.
Here, we describe current and emerging immunomonitoring techniques, especially immunogenomic approaches, and how they are used to predict or monitor immune status in clinical samples (Figure 1). We also describe state-of-the-art computational methods that are being used to quantify immune cell subsets through gene expression data generated from microarray, the nCounter technique, regular RNA-seq, and single-cell level RNA-seq. This review is intended to summarize the development and application of immunomonitoring approaches, and discuss the challenges that must be addressed to accurately quantify the dynamic changes in immune cell composition using bulk RNA-seq data from clinical samples. Other challenges facing the field include incorporating immunotherapy into adjuvant and neoadjuvant cancer therapy; using immunomonitoring data to refine dose, schedule, and duration of treatment; and developing novel surrogate endpoints that accurately capture overall survival benefit early in the treatment course.
Classical Immune Monitoring Techniques
Several classical immunoassays routinely performed for disease diagnosis is discussed below and summarized in Figure 1. Though several new approaches to immunomonitoring are currently under development, recent advances in traditional immunoassays such as enzyme-linked immunosorbent assays (ELISA) and real-time quantitative PCR (RT-PCR) also have huge potential for expanding the breadth of immune monitoring.
Elisa
ELISA is an antibody-based assay that is used to quantify protein, peptides, and antibodies. An ELISA is typically performed with an antigen immobilized on a solid surface while a detection enzyme can be linked directly to the matching antibody (primary antibody) over the surface or introduced through a secondary antibody that recognizes the primary antibody. ELISA is cost-effective and easy to perform, and it is widely used in disease diagnosis throughout the world, especially for infectious diseases such as chronic hepatitis C and chronic hepatitis B, as well as certain cancers, including hepatocellular carcinoma 9, 10. Although classic ELISA is a simple test that measures a single analyte with limited sensitivity, to date ELISA techniques have been independently developed by many laboratories to detect levels of various cytokines and other immune factors in blood, body fluid, and cell culture supernatants 11, 12. For instance, modified ELISA tests with high sensitivity and selectivity have been developed to detect monoclonal antibodies (such as bevacizumab) that are used for cancer immunotherapy 13, 14. Newer bead-based immunoassays utilize the same principle as ELISA but allow simultaneous detection of multiple analytes 15. These multiplex ELISA assays have been validated for in vitro diagnosis and monitoring the early effects of immunotherapy based on plasma markers in pericarditis patients 16. In 2016, a test called a digital ELISA that uses high-affinity autoantibodies was developed for the detection of cytokines at attomolar concentrations, about thousand-fold more sensitive than a traditional ELISA 17. Digital ELISA is able to detect cytokines expressed at very low levels in plasma (such as CXCL10) as markers of specific source cells (such as CXCR3+ T cells) 18, 19, 20.
Elispot
The Enzyme-Linked Immunospot (ELISPOT) assay measures the enzymatic activity of antigen-specific cells with antigen(s) of interest. Unlike ELISA or other techniques, ELISPOT assays detect the secreted analytes (mostly cytokines) directly around the secreting cell, rather than after those analytes are diluted in the supernatant, absorbed by receptors of adjacent cells, or degraded by proteases 21. Although the ELISPOT was initially developed to detect antibody-releasing cells, it has been adapted to quantify cytokine-secreting cells and has become the most sensitive approach to directly monitor and evaluate T cell- and B cell-mediated cellular immunity in vitro 22, 23. Today, ELISPOT is one of the most common immunoassays used in clinical trials of immunotherapies and other immune studies around the world 24. Given that traditional ELISPOT requires proper immune cell preparation and long incubation time, a number of protocol enhancements have been introduced over the last decade to modify and improve ELISPOT and facilitate its application in clinical practice 25, 26, 27. For example, a recent study reported the successful development of a multi-color ELISPOT assay—the FLUOROSPOT—that uses an automatic image acquisition unit to generate colorful fluorescent spots that can be quantified 28.
Flow Cytometry
Clinical immunology analysis performed by ELISA and ELISPOT is very much complemented by flow cytometry techniques. Flow cytometry is a powerful tool to analyze multiple analytes via a variety of parameters on an individual cell basis 29. Recently, development of monoclonal antibodies and immunofluorescence analysis as incorporated in flow cytometry provided many highly sensitive approaches to analyze immune response and change of human mononuclear cell subsets. Compared to other techniques, flow cytometry provides more detailed information on complex immune cell populations by recognizing and counting cells via specific cell surface markers. Flow cytometry is one of the most commonly used methods to measure immune response at single-cell level resolution 30. Recognition of cell surface proteins can identify distinct cell subsets as well as their activation status and many other functional immune features, such as response to a vaccine or immunotherapy 31. Flow cytometry is highly adaptable, not only for the evaluation of immune responses, but also for investigation of cell functions, such as cell apoptosis.
As the demand for immune monitoring technologies has grown, however, the disadvantages of flow cytometry have become apparent, including lack of reproducibility for complex panels, the need for expensive instrumentation and high standards for technicians and maintenance, and limited information observed from on intra-cellular events 32, 33. The increasing number of fluorophores and antibodies available have provided technical innovations in flow cytometry to fit the need from an increasing number of researchers 34. A classic example is flow cytometry was developed and continually modified to evaluate the frequencies of peripheral blood CD4+ and CD8+ T cells in whole blood in patients with cancer, autoimmune diseases and infectious diseases since 1980s 35, 36, 37. The quantity and quality of tumor antigen-specific effector CD4+ and CD8+ T cells are essential markers for monitoring cancer immunotherapies, and flow cytometry is ubiquitously used for immune monitoring in preclinical tumor immunology and in cancer immunotherapy trials 29. In a phase II clinical trial of anti-PD-1 treatment and radiotherapy, flow cytometry was used to identify peripheral blood biomarkers in mononuclear cells and serum samples 38. More recently, a modified flow cytometry was used for advanced analysis of T cells (including regulatory T cells), B cells, natural killer (NK) cells, monocytes, and dendritic cells 39. Improvements in the standardization of flow cytometry labeling and operating protocols has reduced the whole blood sample size required for flow cytometry to less than 2 ml. These “deep immunophenotyping” flow cytometry panels constitute a powerful tool for immune monitoring in autoimmune and cancer clinical trials 39.
RT-PCR
RT-PCR is a powerful tool for quantifying and comparing expression profiles of genes of interest. In RT-PCR (or quantitative PCR, qPCR) techniques, fluorescent signals from the PCR reaction are monitored and tracked over time, then the reaction is stopped before it reaches a plateau. The rate of PCR amplification is then used to quantify the starting amount of genomic material. Because of its high sensitivity, RT-PCR has become the gold standard technique for quantifying cDNA, gDNA, and RNA transcripts (with quantitative reverse transcription PCR) in cultured or primary cells, body fluids, or tissue, as markers of immune response 40. This is the most common and typical immunomonitoring approach used to evaluate changes in human immune system status in response to infection, vaccination, or immunotherapies. For instance, in a phase I clinical trial in patients with acute lymphoblastic leukemia, RT-PCR was used to quantify the copy number of the CAR transgene in CAR-T cells 41. In other pre-clinical studies and immunological research, RT-PCR is commonly used to measure cytokine mRNA levels in inflammation as a way to dissect the early steps of the immune response 42, 43, 44, 45, 46. Similar to ELISA and ELISPOT, highly multiplexed RT-PCR has been developed that allows simultaneous detection and reliable measurement of highly polymorphic target antigens and pathogens in clinical samples 47, 48. Multiplexed RT-PCR is superior to ELISA in terms of diagnostic sensitivity and specificity, and has the advantage of being able to discriminate between species 49.
Despite its popularity, the data generated from RT-PCR can be highly variable and may not be reproducible without appropriate validation tests. Digital PCR is a new approach that offers the possibility of absolute quantification. Digital PCR has been applied to the detection of circulating cell-free DNA, which may serve as a unique tumor marker in certain virus-associated malignancies. Digital droplet PCR was designed to genotype and quantify circulating HPV DNA in patients with HPV16- or HPV18-positive metastatic cervical cancer 50. Moreover, circulating tumor DNA (ctDNA) is one of the best-known indicators of response to anti-PD1 and somatic alterations in ctDNA can be quantified by digital PCR to reveal the gene mutation status of the tumor tissue 51, 52. As measured by digital PCR, if a significant decrease in the amount of ctDNA relative to the baseline level is observed after treatment, it indicates a lack of clinical benefit under anti-PD1 therapy 53, 54, 55, 56. Digital PCR is also an accurate method to assess differentially methylated genomic signatures of immune cells, a marker of immune cell activation in various disease states 57.
Emerging Immunomonitoring Techniques
For decades, immunologists have relied heavily on ELISA and flow cytometry to study human immune responses with low power and minor efficiency. Over the past dozen years, mass spectrometry, mass cytometry, and genomics and transcriptomics approaches have offered higher throughput, and more detailed glimpses into the immune responses in diseases such as cancer and infections, and response to therapies. The recent development of several key techniques is briefly introduced below and summarized in Figure 1.
Figure 1.Overview of classical and emerging immunomonitoring approaches
Mass Spectrometry
Mass spectrometry (MS) accurately measures the mass-to-charge ratio of ions to identify and quantify different molecules within a single sample in an unbiased manner. MS-based proteomics makes it possible to evaluate protein expression, subcellular localization, post-translational modifications, and interactions in immune cells upon activation 58. Many MS-based methods have been developed to study specific targets of the immune response 59. For instance, an MS assay was developed to detect serum prostate-specific antigen (PSA) protein levels, with results correlating well with those from ELISA immunoassays 60, 61. MS is also used to rapidly and reliably quantify catecholamines in peripheral blood mononuclear cells (PBMCs) with significantly improved sensitivity and selectivity as compared to classical immunomonitoring techniques. MS was used to improve understanding of the network between the nervous and immune systems and its dysfunction in various autoimmune and neurological diseases 62, 63, 64. MS-based imaging techniques have been used to visualize the spatial distribution of molecules to analyze biomarkers, metabolites, peptides, and proteins by their molecular masses. MS-based imaging was successfully used to directly isolate and identify special outer membrane vesicles secreted by bacteria such as Pseudomonas aeruginosa , which is the most common cause of lung infection in genetic disease cystic fibrosis and major cause of morbidity and mortality 65. In immunology research, mass spectrometric immunoassay (MSIA) was developed as a rapid, top-down method with high sensitivity and precision for quantification of many clinical analytes 66, 67. A newer and more accurate variation of MSIA, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS), has been used to identify various immune cell subsets and associated modifications in cell activity 68. Reliable insulin-like growth factor 1 (IGF1) quantification in human plasma in the range of 10-1000 ng/mL was achieved by employing MALDI-TOF MS, producing results that were highly correlated with a conventional IGF1 immunoassay 69. The major limitation of current MS and MS-based techniques is the need for large amounts of sample.
Mass Cytometry
Technological progress in the realm of single-cell analysis has been a major driver of clinical immunology research and immunotherapies. Mass cytometry (or cytometry by time-of-flight, CyTOF) is a newer form of flow cytometry in which antibodies are labeled with heavy metal ion tags rather than fluorochromes 70, 71. Compared to conventional flow cytometry, CyTOF allows for single-cell, high-speed analysis and provides simultaneous interrogation of more than 50 metal probes targeting cytokines and transcription factors 3, 72. Mass cytometry has enabled comprehensive immunomonitoring and functional assessments of the complex changes in the immune system in pre-clinical and clinical studies 73. Mass cytometry was used to recognize different subsets of NK T cells in many diseases 74. CD4+ and CD8+ T-cells as well as their ratios can be analyzed by mass cytometry, for example, in the study of PBMCs from HIV patients undergoing antiretroviral therapy 75. The mass cytometry technique also allows identification of novel features in recognized immune cell subsets or new cell populations during disease development. Recently, mass cytometry was performed to identify a distinct monocyte cytokine signature shared by clinically heterogeneous pediatric patients with systemic lupus erythematosus. This signature was observed in CD14+ monocytes, with concomitant increased levels of monocyte chemoattractant protein-1 (MCP1), macrophage inflammatory protein-1beta (MIP-1beta), and interleukin-1 receptor antagonist (IL-1RA) 76. Similarly, new regulatory myeloid cell phenotypes and their clinical impact were recently revealed by mass cytometry 77. In another study, mass cytometry was used to interrogate 30 cell biomarkers at the single-cell level to dissect the effects of graphene oxide T-cell and monocyte activation nanomaterials on distinct immune cells 78. These studies hint at the potential applications of single-cell mass cytometry to the clinical analysis pipeline and diagnostic protocols. Though the use of mass cytometry for studying cancer in the context of cancer immunotherapy has been highlighted in several recent reviews, it does have some technical limitations, including reliance on antibody specificity and quality, expertise required in sample processing and data analysis, the possibility of heavy metal contamination, and challenges regarding reproducibility and high cost 79, 80, 81. Despite many advantages over traditional immune monitoring techniques are showed in mass cytometry, mass cytometry has some technical limitations including limitations in sampling efficiency, slow acquisition speed, challenge of reproducibility, need of high quality antibody, and limited clinical accessibility and feasibility. Although the technique is rapidly developing, the instrument expense and running costs of mass cytometry for now are still much higher compared to conventional immune monitoring techniques such as flow cytometry.
Gene Expression Based and Immunogenomic Approaches
Until recently, the major immune components could only be analyzed through antibody-based protein arrays, with PCR as the only option for quantifying nucleic acids. Recent advances in genomic technologies have broadened the possibilities for using gene expression as a powerful marker of immune system function, where multiple responding cells and cytokines in the tissue or blood could be measured simultaneously. Several methods, including those introduced above, can monitor changes in immune cells to reveal their distinct functionality in health and disease 82. These techniques mostly rely on the quality of antibody, not high throughput-compatible and are limited to tracking a small number of cell subsets or require fresh, live cell samples. In contrast, gene expression profiling of heterogeneous cell samples can detect distinct cell types within populations with high statistical power and strong sensitivity, even in samples with a limited number of cells 83. The deconvolution of cell-specific gene expression signals can yield dynamic estimates of cell population proportions 84. Recent computational algorithms offer parallel and powerful approaches for inferring changes in immune cell quantities from gene expression data 85. However, many profiling studies have primarily focused on mRNA in tissue samples, and therefore required high cellular integrity and minimal tissue destruction. Compared to tissue samples, blood cell samples are easy to access and can be collected before and after treatment for RNA extraction and transcriptome analysis 86, 87. In particular, gene expression microarray and NGS approaches (such as RNA-seq) are the most successful technologies emerging from the work of the Human Genome Project. Application of these approaches to immunomonitoring is discussed in more detail in Section 5 below.
The Current and Urgent Need for Immunomonitoring
Given significant progresses have taken place in the field of cancer immunotherapeutics, immunomonitoring during immunotherapy have become an emerging need as more and more immunotherapies are made available for disease including but not limited to cancer. The overall goal of immunomonitoring is to screen the “responders” and thus determine which strategy would benefit. Taking cancer as an example, on one hand cancer-induced abnormalities in the immune system suppresses the cancer immune surveillance, on the other hand cancer cells also limit the effect of cancer immunotherapy by regulating immune network and cell functions. The dynamic and complex immune status of individual determined that monitoring and manipulating the immune status in a timely manner is a key process during immunotherapy. There are two types of immunotherapies: active immunotherapy activates the immune system of the host to attack tumor cells by targeting tumor antigens; passive immunotherapy enhances existing immune responses initiated by external antibodies or other immune components such as checkpoint inhibitors, adoptive transfer of lymphocytes, macrophages, or cytokines 88. Currently, several immune checkpoint blockade therapies have been approved by the FDA for the treatment of a broad range of cancer types. Stimulatory and inhibitory cell surface proteins are manipulated in cancer immunotherapies, thereby regulating immune cell function and interaction of immune cells with tumor cells 89. In addition to immune checkpoint blockers, currently available passive immunotherapies have been developed to treat infectious diseases 90. Current therapeutic regimens for various infectious diseases involve the prolonged use of antibiotics with potential side effects and may lead to bacterial resistance. Immunotherapy has become a powerful tool to combat the risks associated with overuse of antibiotics. The emergence of pathogen-specific monoclonal antibodies has breathed new life into the immunotherapy field as researchers seek non-antibiotic interventions for respiratory infections caused by multi-drug resistant bacteria 91. A phase III study showed that the immunomodulator immunoxel and recombinant human interleukin-2 could reduce pulmonary tuberculosis-associated inflammation, providing an affordable and fast-acting immunotherapeutic intervention to supplement conventional chemotherapy 92, 93.
In patients with either cancer or infectious disease treated by immunotherapies, the change in immune cells and immunological pathways in response to immunotherapies need to be assessed and linked to clinical outcomes 94, 95. Compared to active immunotherapy, passive immunotherapy uses adaptive materials that modulate the immune system in an unpredictable manner. Despite of the remarkable progress of clinical applications of checkpoint blockade, the efficacy of these immunotherapies are currently limited to individual patients, tumor types, and indications. There is a need for more effective and novel immunomonitoring approaches that can be used to predict response and better select responder patients 96. Using biomarkers in immune monitoring for prediction of treatment efficacy, immune tolerance, and disease progression has to potential to improve therapeutic outcomes by matching treatments to patients 97. Thus, there is an urgent need to identify potential biomarkers, and develop their accompanying assays, for timely and accurate monitoring of treatment response in patients receiving immunotherapies.
Immunogenomic Approaches to Monitor the Immune Landscape
Genome-wide gene expression data derived from bulk tumor tissue samples has been used to study immune cell infiltration within the tumor microenvironment and define molecular subtypes of many different types of cancers 98. These data were mostly generated from immunogenomic platforms such as immune gene expression microarrays, multiplex flow cytometry panels, and RNA-seq, providing a more holistic picture of the different parts of the immune system from a system immunology perspective 99, 100. Although flow cytometry is capable of measuring more than 50 immune cell populations to comprehensively identify the immune cell composition in cancer tissue 101, 102, a couple of major limitations hinder the routine use of flow cytometry or mass cytometry methods in the determination of immune cell compositions in large-scale samples. These limitations include 1) high cost and requirement of large volume of samples; 2) loss of cell in sample processing and lack availability to analyze the dead cells; and 3) some types of immune cells have no suitable cell surface markers or no good quality antibodies 103. RNA-seq is a more powerful tool to profile immune cells and assess detailed information like non-coding RNA and splice variants that regular gene expression microarray is not able to obtain. However, RNA-seq requires lengthy analytical approaches and long data processing times, which limits the application of RNA-seq in studies of large sample sizes in clinical setting. Nevertheless, a few gene expression-based computational methods have been developed for immune cell composition analysis in both tissue and blood samples 82. A few typical studies are discussed below and a full summary of these studies is provided in Table 1.
In a systems biology study published in 2014, a computational method named digital cell quantification (DCQ) was used to study global immune cell dynamics in mouse lungs at 10 time points during 7 days of flu infection 104. This method combines genome-wide gene expression data with an immune cell compendium to infer in vivo changes in the quantities of 213 immune cell subpopulations. Dramatic changes in quantities of 70 immune cell types were observed, including various innate, adaptive, and progenitor immune cells. In 2015, a robust approach based on unique molecular identifiers (UMI), called UMI-based quantification, was reported to identify Ag-specific lymphocyte subpopulations from several hundred to several thousand lymphocytes, preserving qualitative and quantitative information on clonal composition of the sample. This data analysis provided accurate counting of starting molecules in high-throughput sequencing applications 105. The study demonstrated that gene expression-based computational methods are powerful tools for analysis following normalized, error-free sequencing in an application for qualified analysis of Ag receptors in minor lymphocyte subsets. Also in 2015, a nu-support vector regression (SVR) based method termed CIBERSORT (http://cibersort.stanford.edu/) showed significant advantages in enumeration of immune cell subsets in RNA mixtures from fresh, frozen, and fixed tissues, including solid tumors 84. That study inferred the proportions of 22 subsets of immune cells using a well-designed signature matrix optimized for human cancer deconvolution to characterize the cell composition of complex tissues from their gene expression profiles. The same approach was used to investigate the associations between immune cell type, survival, and sensitivity of breast cancer to chemotherapy 106. In 2016, another similar method, MCP-counter, identified immune infiltrates across human healthy tissues and non-hematopoietic human tumors as well as microenvironment-based patient stratifications associated with overall survival in lung adenocarcinoma and colorectal and breast cancer 85. In 2017, a scientist from NanoString reported on an analysis method that identified a list of 60 high confidence marker genes representing 14 immune cell populations to create a cell type score 83. The gene signature-calculated cell type scores were consistent with flow cytometry and immunohistochemistry (IHC) findings. A similar method named ImSig demonstrated how cell-specific gene markers can be used for the quantitative estimation of immune cell content of melanoma and nontumor tissue samples. The ImSig could also identify immune cells with the use of single-cell RNA-seq, and use the results to assign melanoma patients into prognostic subgroups 107. A full summary of a standard workflow used in these studies is showed in Figure 2.
Figure 2.Workflow of immunogenomics based cell subset composition analysis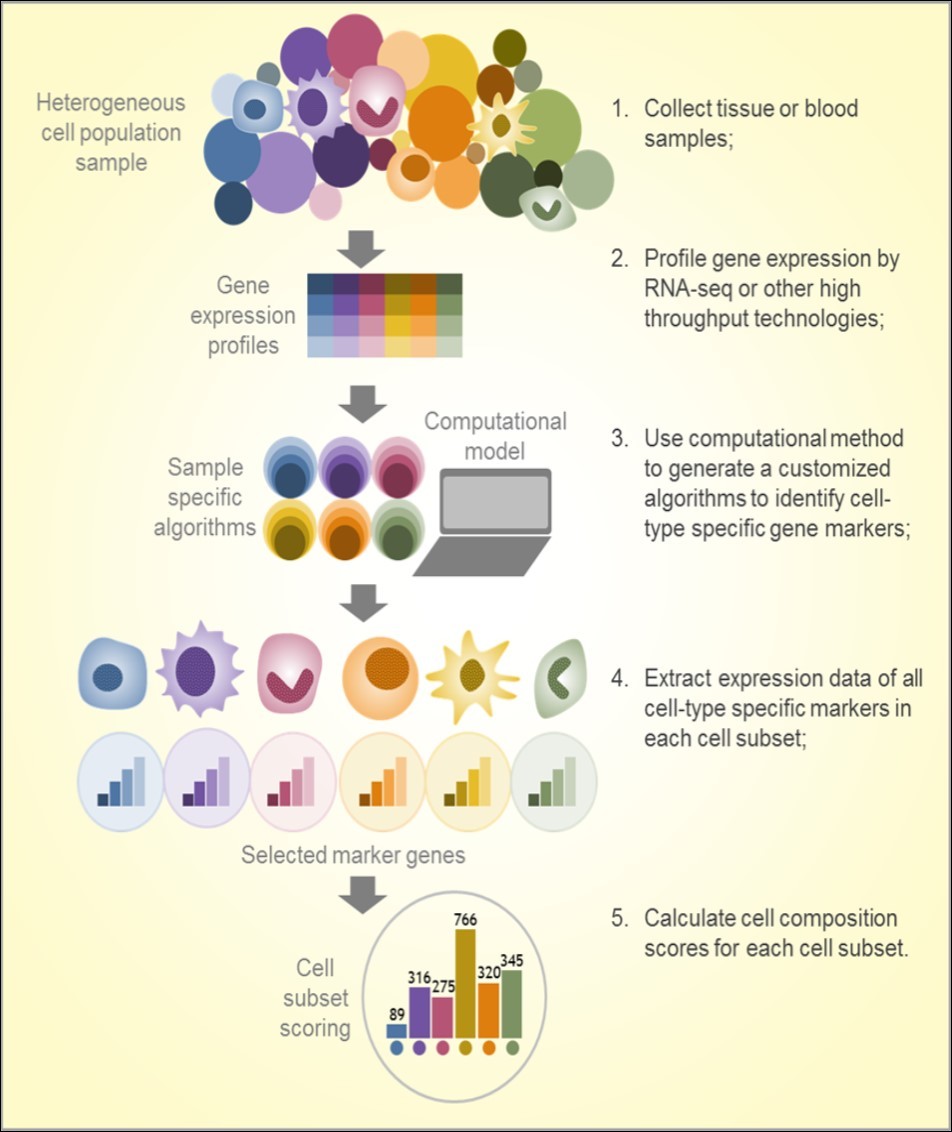
Taken together, a number of immunogenomic studies have demonstrated the utility of computational approaches to analyze immune cell transcriptome signatures, evaluate immune cell subset composition in tissue or PBMCs, and investigate the tumor microenvironment (Figure 2)8. Most of these studies were performed in samples from patients with cancer, but their findings imply great potential for applications in other diseases that may also need immunomonitoring. Knowing the specific immune cell composition in blood cells may provide clues regarding the immune response to antibiotic therapy and immunotherapy in patients with cancer and infectious diseases 98, 108. Lists of marker genes have been (and continue to be) identified and rigorously tested in large and independent samples. The majority of these gene signatures were obtained from microarray data, though some were from RNA-seq, including single-cell RNA-seq (Table 1), suggesting both the intratumoral and peripheral immune cell landscape could be broadly assessed in immunogenomic studies. Due to the computational simplicity and utility of clinical samples, these approaches may be useful in future immunological research and clinical trials to understand how therapeutic interventions shape the local immune landscape in tumor cells and immune cells.
Table 1. Studies of computational approaches used in cell subset composition analysis| Approach name | Application | Sample | Data source | Year | Reference | |||
| Digital cell quantification (DCQ) | Study of global immune cell dynamics in mouse lungs at 10 time points | Lung tissue (mice with flu) | Microarray | 2014 | 104 | |||
| Unique molecular identifiers (UMI)-based quantification | Sorting and profiling specific lymphocyte subpopulations from blood cells | Human PBMCs | PCR and sequencing | 2015 | 105 | |||
| CIBERSORT | Enumerating immune cell subsets in several cancer tissues | Cancer tissue (human) | Microarray | 2015 | 84 | |||
| CIBERSORT | Correlation of immune cell type and survival and response to chemotherapy in breast cancer | Breast cancer tissue (human) | Published gene expression data | 2016 | 106 | |||
| Microenvironment cell populations (MCP)-counter | Quantification of the absolute abundance of immune and stromal cell populations in heterogeneous cancer tissues | Cancer tissue (human) | Transcriptomic data | 2016 | 85 | |||
| Immune gene signatures | Identification distinct immune-enriched gene signatures in tumor-infiltrating leukocytes and their prognostic implications in several solid tumor tissues | Cancer tissue (human) | Microarray and flow cytometry | 2016 | 109 | |||
| Immune cell subset analysis (NanoString) | Evaluation of immune cell populations in the tumor microenvironment | Cancer tissue (human) | nCounter gene expression data | 2017 | 83 | |||
| ImmuCC | Inferring relative compositions of 25 immune cell types in mouse tissues | Tissue (mice) | Microarray | 2017 | 103 | |||
| Reference gene expression profiles (RGEPs) | Determining cellular composition of solid tumors | PBMCs and cancer tissue (human) | Single-cell RNA-seq | 2017 | 110 | |||
| eTumorType | Non-invasive diagnosis to determine cancer types based on CNVs of CTCs and cfDNAs | Cancer tissue (human) | Microarray (SVP) data from The Cancer Genome Atlas (TCGA) database | 2017 | 111 | |||
| ImSig | Quantification of immune cell content in tumor and non-tumor tissue samples | Cancer tissue (human) | Transcriptomic data | 2018 | 107 | |||
| MySort | Identification of immune cell types from blood biopsies | Human PBMCs and immune cells | Published microarray data | 2018 | 112 | |||
| Seq-ImmuCC | Assessing the relative proportions of 10 major immune cells in mouse tissues from RNA-seq data | Tissue (mice) | RNA-seq | 2018 | 113 | |||
| MCP-counter and RNA deconvolution method | Examining the immune cell subset composition in the tumor microenvironment of colorectal cancer | Colorectal cancers tissue (human) | Whole-genome sequencing and RNA-seq | 2018 | 114 | |||
| TSNet | To identify novel co-expression modules and hub structure specific to tumor cells. | Cancers tissue (human) | Ovarian cancer RNAseq data | 2018 | 115 | |||
Conclusion
Overall, computational analysis utilizing gene expression data from preclinical/clinical samples is one of the most effective approaches to assessing and characterizing molecular-level changes in the immune landscapes from large groups of patients undergoing therapy for various diseases and conditions. Future research should address the association between immune cell responses to therapies measured using these approaches and other clinical parameters of therapeutic response. Immunogenomics may also be applied to the development and assessment of targeted therapies complementary to existing immunotherapies. Though many limitations and difficulties remain in developing immunogenomic approaches to monitor the immune landscape, recent advances and their application to clinical practice may help guide patient selection and treatment modification to optimize clinical responses and outcomes.
Funding
This work was supported from National Natural Science Foundation of China (No. 81672627 to XZ).
References
- 1.Chen J, Xiao-Zhong G, Qi X S. (2017) Clinical Outcomes of Specific Immunotherapy in Advanced Pancreatic Cancer: A Systematic Review and Meta-Analysis. Journal of immunology research. 8282391.
- 2.Emens L A, Ascierto P A, Darcy P K, Demaria S, AMM Eggermont et al. (2017) Cancer immunotherapy: Opportunities and challenges in the rapidly evolving clinical landscape. , Eur J Cancer 81, 116-129.
- 3.Greenplate A R, Johnson D B, Ferrell P B, Irish J M. (2016) Systems immune monitoring in cancer therapy. , Eur J Cancer 61, 77-84.
- 4.Nishino M, Ramaiya N H, Hatabu H, Hodi F S. (2017) Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nature reviews Clinical oncology. 14, 655-668.
- 5.Kohrt H E, Tumeh P C, Benson D, Bhardwaj N, Brody J et al. (2016) Immunodynamics: a cancer immunotherapy trials network review of immune monitoring in immuno-oncology clinical trials. Journal for immunotherapy of cancer. 4-15.
- 6.Boyd S D, Hoh R A, Nadeau K C, Galli S J. (2017) Immune monitoring for precision medicine in allergy and asthma. Current opinion in immunology. 48, 82-91.
- 7.Wargo J A, Reddy S M, Reuben A, Sharma P. (2016) Monitoring immune responses in the tumor microenvironment. Current opinion in immunology. 41, 23-31.
- 8.Finotello F, Trajanoski Z. (2018) Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol Immunother. 67, 1031-1040.
- 9.Usuda S. (2004) [Differentiation of hepatitis B virus genotypes by ELISA using monoclonal antibodies]. Nihon Rinsho. , 62 Suppl 8, 163-166.
- 10.Kim M H, Lee H J, Park S Y, Lee Y S, Suh J T. (2006) . , [Usefulness of Anti-HCV ELISA Test and HCV Reverse Transcriptase-PCR for the Diagnosis of Hepatits C Viral Infection.]. Korean J Lab Med 26, 418-423.
- 11.Pang B, Zhao C, Li L, Song X, Xu K et al. (2018) Development of a low-cost paper-based ELISA method for rapid Escherichia coli O157:H7 detection. Anal Biochem. 542-58.
- 12.Wurzner R, Tedesco F, Garred P, Mollnes T E, Truedsson L et al. (2015) European Union funded project on the development of a whole complement deficiency screening ELISA-A story of success and an exceptional manager: MohamedR.Daha.MolImmunol. 68, 63-66.
- 13.Hamidaddin M A, AlRabiah H, Darwish I A. (2018) Development and validation of generic heterogeneous fluoroimmunoassay for bioanalysis of bevacizumab and cetuximab monoclonal antibodies used for cancer immunotherapy. Talanta. 188, 562-569.
- 14.Suarez I, Salmeron-Garcia A, Cabeza J, Capitan-Vallvey L F, Navas N. (2016) Development and use of specific ELISA methods for quantifying the biological activity of bevacizumab, cetuximab and trastuzumab in stability studies. Journal of chromatography B,Analytical technologies in the biomedical and life sciences.1032:. 155-164.
- 15.Tighe P J, Ryder R R, Todd I, Fairclough L C. (2015) ELISA in the multiplex era: potentials and pitfalls. Proteomics Clin Appl. 9, 406-422.
- 16.Shenje J, Lai R P, Ross I L, Mayosi B M, Wilkinson R J et al. (2018) Effect of prednisolone on inflammatory markers in pericardial tuberculosis: A pilot study. , Int J Cardiol Heart Vasc 18, 104-108.
- 17.Leirs K, Tewari Kumar P, Decrop D, Perez-Ruiz E, Leblebici P et al. (2016) Bioassay Development for Ultrasensitive Detection of Influenza A Nucleoprotein Using Digital ELISA. Anal Chem. 88, 8450-8458.
- 18.Perez-Ruiz E, Decrop D, Ven K, Tripodi L, Leirs K et al. (2018) Digital ELISA for the quantification of attomolar concentrations of Alzheimer's disease biomarker protein Tau in biological samples. Analytica chimica acta. 1015, 74-81.
- 19.ERB Petersen, Olsen D A, Christensen H, Hansen S B, Christensen C et al. (2017) Rhodopsin in plasma from patients with diabetic retinopathy - development and validation of digital ELISA by Single Molecule Array (Simoa) technology. , J Immunol Methods 446, 60-69.
- 20.Blauenfeldt T, Petrone L, Del Nonno F, Baiocchini A, Falasca L et al.Interplay of DDP4 and IP-10 as a Potential Mechanism for Cell Recruitment to Tuberculosis Lesions. Frontiers in immunology 2018; 9:. 1456.
- 21.Slota M, Lim J B, Dang Y, Disis M L. (2011) ELISpot for measuring human immune responses to vaccines. Expert review of vaccines. 10, 299-306.
- 22.Karulin A Y, Caspell R, Dittrich M, Lehmann P V. (2015) Normal Distribution of CD8+ T-Cell-Derived ELISPOT Counts within Replicates Justifies the Reliance on Parametric Statistics for Identifying Positive Responses. Cells. 4, 96-111.
- 23.Fuchs Y F, Jainta G W, Kuhn D, Wilhelm C, Weigelt M et al. (2015) Vagaries of the ELISpot assay: specific detection of antigen responsive cells requires purified CD8(+) T cells and MHC class I expressing antigen presenting cell lines. , Clin Immunol 157, 216-225.
- 24.JDC Lima-Junior, Morgado F N, Conceicao-Silva F. (2017) How Can Elispot Add Information to Improve Knowledge on Tropical Diseases? Cells. 6.
- 25.Kobayashi T, Sato J I, Ikuta K, Kanno R, Nishiyama K et al. (2017) Modification of the HCMV-specific IFN-gamma release test (QuantiFERON-CMV) and a novel proposal for its application. Fukushima journal of medical science. 63, 64-74.
- 26.Horvati K, Bosze S, Gideon H P, Bacsa B, Szabo T G et al. (2016) Population tailored modification of tuberculosis specific interferon-gamma release assay. The Journal of infection. 72, 179-188.
- 27.Santos R, Buying A, Sabri N, Yu J, Gringeri A et al. (2014) Improvement of IFNg ELISPOT Performance Following Overnight Resting of Frozen PBMC Samples Confirmed Through Rigorous Statistical Analysis. Cells. 4, 1-18.
- 28.Karulin A Y, Megyesi Z, Caspell R, Hanson J, Lehmann P V. (2018) Multiplexing T- and B-Cell FLUOROSPOT Assays:. Experimental Validation of the Multi-Color ImmunoSpot((R)) Software Based on Center of Mass Distance Algorithm. Methods Mol Biol 1808, 95-113.
- 29.Santegoets S J, Welters M J, SH van der Burg. (2016) Monitoring of the Immune Dysfunction in Cancer Patients. Vaccines. 4.
- 30.Irish J M, Doxie D B. (2014) High-dimensional single-cell cancer biology. Current topics in microbiology and immunology. 377, 1-21.
- 31.Macchia I, Urbani F, Proietti E. (2013) Immune monitoring in cancer vaccine clinical trials: critical issues of functional flow cytometry-based assays. BioMed research international. 726239.
- 32.Tenorio-Borroto E, Ramirez F R, Speck-Planche A, Cordeiro M N, Luan F et al. (2014) QSPR and flow cytometry analysis (QSPR-FCA): review and new findings on parallel study of multiple interactions of chemical compounds with immune cellular and molecular targets. Current drug metabolism. 15, 414-428.
- 33.Grigore A, Albulescu A, Albulescu R. (2018) Current methods for tumor-associated macrophages investigation. , J Immunoassay Immunochem 39, 119-135.
- 34.Pane M, Allesina S, Amoruso A, Nicola S, Deidda F et al.Flow Cytometry: Evolution of Microbiological Methods for Probiotics Enumeration. Journal of clinical gastroenterology 2018; 52 Suppl 1. Proceedings from the 9th Probiotics, Prebiotics and New Foods, Nutraceuticals and Botanicals for Nutrition & Human and Microbiota Health Meeting, held in Rome, Italy fromSeptember10to12,2017. S41-S45 .
- 35.Niitsu N, Kohri M, Togano T, Nakamine H, Nakamura S et al. (2004) Development of hepatosplenic gammadelta T-cell lymphoma with pancytopenia during early pregnancy: a case report and review of the literature. European journal of haematology. 73, 367-371.
- 36.Kita H, He X S, Gershwin M E. (2003) Application of tetramer technology in studies on autoimmune diseases. Autoimmunity reviews. 2, 43-49.
- 37.Lucey D R, Clerici M, Shearer G M. (1996) Type 1 and type 2 cytokine dysregulation in human infectious, neoplastic, and inflammatory diseases. Clinical microbiology reviews. 9, 532-562.
- 38.K De Wolf, Kruse V, Sundahl N, M van Gele, Chevolet I et al. (2017) A phase II trial of stereotactic body radiotherapy with concurrent anti-PD1 treatment in metastatic melanoma: evaluation of clinical and immunologic response. , Journal of translational medicine 15-21.
- 39.Pitoiset F, Cassard L, K El Soufi, Boselli L, Grivel J et al. (2018) Deep phenotyping of immune cell populations by optimized and standardized flow cytometry analyses. Cytometry Part A : the journal of the International Society for Analytical Cytology. 93, 793-802.
- 40.Stordeur P. (2009) Monitoring the immune response using real-time PCR. , Methods Mol Biol 496, 323-338.
- 41.Tang X Y, Sun Y, Zhang A, Hu G L, Cao W et al. (2016) Third-generation CD28/4-1BB chimeric antigen receptor T cells for chemotherapy relapsed or refractory acute lymphoblastic leukaemia: a non-randomised, open-label phase I trial protocol. , BMJ Open 6, 013904.
- 42.Talei M, Abdi A, Shanebandi D, Jadidi-Niaragh F, Khabazi A et al. (2018) . Interleukin-33 Gene expression and rs1342326 Polymorphism in Behcet's Disease. Immunology letters .
- 43.Zhang G, Liu H B, Zhou L, Cui X Q, Fan X H. (2018) CCL3 participates in the development of rheumatoid arthritis by activating AKT. European review for medical and pharmacological sciences. 22, 6625-6632.
- 44.Braga Diniz JM, Espaladori M C, ESME Souza, LCN Brito, Vieira L Q. (2018) Ribeiro Sobrinho AP. Immunological profile of teeth with inflammatory periapical disease from chronic liver disease patients. International endodontic journal.
- 45.Zhang Y, Li H, Jia X, Zhang X, Xia Y et al. (2009) Increased expression of P2X7 receptor in peripheral blood mononuclear cells correlates with clinical severity and serum levels of Th17-related cytokines in patients with myasthenia gravis. Clinical neurology and neurosurgery. 157, 88-94.
- 46.Qu M M, Liu X N, Liu X G, Feng Q, Liu Y et al. (2017) Cytokine changes in response to TPO receptor agonist treatment in primary immune thrombocytopenia. Cytokine. 92, 110-117.
- 47.Manukyan H, Zagorodnyaya T, Ruttimann R, Manor Y, Bandyopadhyay A et al. (2018) Quantitative multiplex one-step RT-PCR assay for identification and quantitation of Sabin strains of poliovirus in clinical and environmental specimens. , J Virol Methods 259, 74-80.
- 48.Laamiri N, Aouini R, Marnissi B, Ghram A, Hmila I. (2018) A multiplex real-time RT-PCR for simultaneous detection of four most common avian respiratory viruses. , Virology 515, 29-37.
- 49.Ng-Nguyen D, Stevenson M A, Dorny P, Gabriel S, Vo T V et al. (2017) Comparison of a new multiplex real-time PCR with the Kato Katz thick smear and copro-antigen ELISA for the detection and differentiation of Taenia spp. in human stools. PLoS Negl Trop Dis. 11, 0005743.
- 50.Kang Z, Stevanovic S, Hinrichs C S, Cao L. (2017) Circulating Cell-free DNA for Metastatic Cervical Cancer Detection, Genotyping, and Monitoring. Clin Cancer Res. 23, 6856-6862.
- 51.Sefrioui D, Beaussire L, Perdrix A, Clatot F, Michel P et al. (2017) Direct circulating tumor DNA detection from unpurified plasma using a digital PCR platform. Clin Biochem. 50, 963-966.
- 52.Postel M, Roosen A, Laurent-Puig P, Taly V, Wang-Renault S F. (2018) Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: a cancer diagnostic perspective. Expert review of molecular diagnostics. 18, 7-17.
- 53.Herbreteau G, Vallee A, Knol A C, Theoleyre S, Quereux G et al. (2018) Quantitative monitoring of circulating tumor DNA predicts response of cutaneous metastatic melanoma to anti-PD1 immunotherapy. Oncotarget. 9, 25265-25276.
- 54.Ashida A, Sakaizawa K, Uhara H, Okuyama R. (2017) Circulating Tumour DNA for Monitoring Treatment Response to Anti-PD-1 Immunotherapy in Melanoma Patients. Acta dermato-venereologica. 97, 1212-1218.
- 55.Cabel L, Bidard F C, Servois V, Cacheux W, Mariani P et al. (2017) HPV circulating tumor DNA to monitor the efficacy of anti-PD-1 therapy in metastatic squamous cell carcinoma of the anal canal: A case report. , Int J Cancer 141, 1667-1670.
- 56.Cabel L, Riva F, Servois V, Livartowski A, Daniel C et al. (2017) Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Ann Oncol. 28, 1996-2001.
- 57.Wiencke J K, Butler R, Hsuang G, Eliot M, Kim S et al. (2016) The DNA methylation profile of activated human natural killer cells. Epigenetics. 11, 363-380.
- 58.Nyman T A, Lorey M B, Cypryk W, Matikainen S. (2017) Mass spectrometry-based proteomic exploration of the human immune system: focus on the inflammasome, global protein secretion, and T cells. Expert Rev Proteomics. 14, 395-407.
- 59.Purcell A W, Gorman J J. (2004) Immunoproteomics: Mass spectrometry-based methods to study the targets of the immune response. Molecular & cellular proteomics : MCP. 3, 193-208.
- 60.Klee E W, Bondar O P, Goodmanson M K, Trushin S A, Bergstralh E J et al. (2014) Serum concentrations of prostate-specific antigen measured using immune extraction, trypsin digestion, and tandem mass spectrometry quantification of LSEPAELTDAVK peptide. , Archives of pathology & laboratory medicine 138, 1381-1386.
- 61.Klee E W, Bondar O P, Goodmanson M K, Trushin S A, Singh R J et al. (2014) Mass spectrometry measurements of prostate-specific antigen (PSA) peptides derived from immune-extracted PSA provide a potential strategy for harmonizing immunoassay differences. , Am J Clin Pathol 141, 527-533.
- 62.Li X S, Li S, Kellermann G. (2016) An integrated liquid chromatography-tandem mass spectrometry approach for the ultra-sensitive determination of catecholamines in human peripheral blood mononuclear cells to assess neural-immune communication. , Journal of chromatography A 1449, 54-61.
- 63.Giangrande C, Auberger N, Rentier C, Papini A M, Mallet J M et al. (2016) Multi-Stage Mass Spectrometry Analysis of Sugar-Conjugated beta-Turn Structures to be Used as Probes in Autoimmune Diseases. , Journal of the American Society for 27, 735-747.
- 64.Guo D, Gu P, Liu Z, Tang K, Du Y et al. (2015) Proteomic analysis of rat plasma with experimental autoimmune uveitis based on label-free liquid chromatography-tandem mass spectrometry (LC-MS/MS). Journal of chromatography B,Analytical technologies in the biomedical and life sciences.976-977:. 84-90.
- 65.Lahiri P, Ghosh D. (2017) Single-Step Capture and Targeted Metabolomics of Alkyl-Quinolones in Outer Membrane Vesicles of Pseudomonas aeruginosa. , Methods Mol Biol 1609, 171-184.
- 66.Yassine H, Borges C R, Schaab M R, Billheimer D, Stump C et al. (2013) Mass spectrometric immunoassay and MRM as targeted MS-based quantitative approaches in biomarker development: potential applications to cardiovascular disease and diabetes. Proteomics Clin Appl. 7, 528-540.
- 67.Chin C F, Ler L W, Choong Y S, Ong E B, Ismail A et al. (2016) Application of streptavidin mass spectrometric immunoassay tips for immunoaffinity based antibody phage display panning. , Journal of microbiological methods 120, 6-14.
- 68.Ouedraogo R, Textoris J, Daumas A, Capo C, Mege J L. (2013) Whole-cell MALDI-TOF mass spectrometry: a tool for immune cell analysis and characterization. Methods Mol Biol. 1061, 197-209.
- 69.Klont F, Ten Hacken NHT, Horvatovich P, SJL Bakker, Bischoff R. (2017) Assuring Consistent Performance of an Insulin-Like Growth Factor 1 MALDImmunoassay by Monitoring Measurement Quality Indicators. Anal Chem. 89, 6188-6195.
- 70.Chen G, Weng N P. (2012) Analyzing the phenotypic and functional complexity of lymphocytes using CyTOF (cytometry by time-of-flight). Cellular & molecular immunology. 9, 322-323.
- 71.Cheung R K, Utz P J. (2011) Screening:CyTOF-the next generation of cell detection. , Nature reviews Rheumatology 7, 502-503.
- 72.Amir E D, Guo Mayovska O, Rahman A H. (2018) Average Overlap Frequency: A simple metric to evaluate staining quality and community identification in high dimensional mass cytometry experiments. J Immunol Methods.453:.
- 73.Chattopadhyay P K, Gierahn T M, Roederer M, Love J C. (2014) Single-cell technologies for monitoring immune systems. Nature immunology. 15, 128-135.
- 74.Kumar V, Delovitch T L. (2014) Different subsets of natural killer T cells may vary in their roles in health and disease. , Immunology 142, 321-336.
- 75.Serrano-Villar S, Sainz T, Ma Z M, Utay N S, Chun T W et al. (2016) Effects of Combined CCR5/Integrase Inhibitors-Based Regimen on Mucosal Immunity in HIV-Infected Patients Naive to Antiretroviral Therapy: A Pilot Randomized Trial. PLoS pathogens. 12-1005381.
- 76.O'Gorman W E, Kong D S, Balboni I M, Rudra P, Bolen C R et al. (2017) Mass cytometry identifies a distinct monocyte cytokine signature shared by clinically heterogeneous pediatric SLE patients. , Journal of autoimmunity
- 77.Roussel M, Irish J M, Menard C, Lhomme F, Tarte K et al. (2017) Regulatory myeloid cells: an underexplored continent in B-cell lymphomas. Cancer Immunol Immunother. 66, 1103-1111.
- 78.Orecchioni M, Bedognetti D, Newman L, Fuoco C, Spada F et al. (2017) Single-cell mass cytometry and transcriptome profiling reveal the impact of graphene on human immune cells. Nature communications. 8, 1109.
- 79.Simoni Y, MHY Chng, Li S, Fehlings M, Newell E W. (2018) Mass cytometry: a powerful tool for dissecting the immune landscape. Current opinion in immunology. 51, 187-196.
- 80.Behbehani G K. (2017) Applications of Mass Cytometry in Clinical Medicine: The Promise and Perils of Clinical CyTOF. Clinics in laboratory medicine. 37, 945-964.
- 81.Newell E W, Cheng Y. (2016) Mass cytometry: blessed with the curse of dimensionality. Nature immunology. 17, 890-895.
- 82.Lyons Y A, Wu S Y, Overwijk W W, Baggerly K A, Sood A K. (2017) Immune cell profiling in cancer: molecular approaches to cell-specific identification. npj Precision Oncology. 1, 26.
- 83.Danaher P, Warren S, Dennis L, D'Amico L, White A et al. (2017) Gene expression markers of Tumor Infiltrating Leukocytes. , J Immunother Cancer 5, 18.
- 84.Newman A M, Liu C L, Green M R, Gentles A J, Feng W et al. (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 12, 453-457.
- 85.Becht E, Giraldo N A, Lacroix L, Buttard B, Elarouci N et al. (2016) Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 17-218.
- 86.Furman D, Davis M M. (2015) New approaches to understanding the immune response to vaccination and infection. 33, 5271-5281.
- 87.Haralambieva I H, Ovsyannikova I G, Pankratz V S, Kennedy R B, Jacobson R M et al. (2013) The genetic basis for interindividual immune response variation to measles vaccine: new understanding and new vaccine approaches. Expert review of vaccines. 12, 57-70.
- 88.Marabelle A, Gray J. (2015) Tumor-targeted and immune-targeted monoclonal antibodies: Going from passive to active immunotherapy. Pediatric blood & cancer. 62, 1317-1325.
- 90.Manohar A, Ahuja J, Crane J K. (2015) Immunotherapy for Infectious Diseases: Past, Present, and Future. Immunological investigations. 44, 731-737.
- 91.Babb R, Pirofski L A. (2017) Help is on the way: Monoclonal antibody therapy for multi-drug resistant bacteria. Virulence. 8, 1055-1058.
- 92.Batbold U, Butov D O, Kutsyna G A, Damdinpurev N, Grinishina E A et al. (2017) Double-blind,placebo-controlled,1: 1randomized Phase III clinical trial of Immunoxel honey lozenges as an adjunct immunotherapy in 269 patients with pulmonary tuberculosis. , Immunotherapy 9, 13-24.
- 93.Zhang R, Xi X, Wang C, Pan Y, Ge C et al. (2018) Therapeutic effects of recombinant human interleukin 2 as adjunctive immunotherapy against tuberculosis: A systematic review and meta-analysis. PLoS One. 13-0201025.
- 94.Anciaux M, Vandeputte C, Hendlisz A. (2017) Tackling immunomonitoring in gastrointestinal cancer. Current opinion in oncology. 29, 296-305.
- 95.Menezes E G, Coelho-Dos-Reis J G, Cardoso L M, Lopes-Ribeiro A, Jonathan-Goncalves J et al. (2017) Strategies for serum chemokine/cytokine assessment as biomarkers of therapeutic response in HCV patients as a prototype to monitor immunotherapy of infectious diseases.AntiviralResearch. 141.
- 96.Byun D J, Wolchok J D, Rosenberg L M, Girotra M. (2017) Cancer immunotherapy - immune checkpoint blockade and associated endocrinopathies. Nature reviews Endocrinology. 13, 195-207.
- 97.Aptsiauri N, Jewett A, Hurwitz A A, Shurin M R, Umansky V. (2016) Redefining cancer immunotherapy-optimization, personalization, and new predictive biomarkers:. 4th Cancer Immunotherapy and Immunomonitoring (CITIM) meeting,April27-30,2015,Ljubljana, Slovenia. Cancer Immunol Immunother 65, 875-883.
- 98.Thorsson V, Gibbs D L, Brown S D, Wolf D, Bortone D S et al. (2018) The Immune Landscape of Cancer. Immunity.48: 812-830.
- 99.Rabinovich G A, Conejo-Garcia J R. (2016) Shaping the Immune Landscape in Cancer by Galectin-Driven Regulatory Pathways. , Journal of molecular biology 428, 3266-3281.
- 100.Bindea G, Mlecnik B, Angell H K, Galon J. (2014) The immune landscape of human tumors: Implications for cancer immunotherapy. Oncoimmunology. 3, 27456.
- 101.Teixido C, Rosell R.Neutrophils dominate the immune landscape of non-small cell lung cancer. , Journal of thoracic disease 2017, 468-469.
- 102.Kargl J, Busch S E, Yang G H, Kim K H, Hanke M L et al. (2017) Neutrophils dominate the immune cell composition in non-small cell lung cancer. Nature communications. 8, 14381.
- 103.Chen Z, Huang A, Sun J, Jiang T, Qin F X et al. (2017) Inference of immune cell composition on the expression profiles of mouse tissue. Sci Rep. 7, 40508.
- 104.Altboum Z, Steuerman Y, David E, Barnett-Itzhaki Z, Valadarsky L et al. (2014) Digital cell quantification identifies global immune cell dynamics during influenza infection. Mol Syst Biol. 10-720.
- 105.Egorov E S, Merzlyak E M, Shelenkov A A, Britanova O V, Sharonov G V et al. (2015) Quantitative profiling of immune repertoires for minor lymphocyte counts using unique molecular identifiers. , J Immunol 194, 6155-6163.
- 106.Ali H R, Chlon L, Pharoah P D, Markowetz F, Caldas C. (2016) Patterns of Immune Infiltration in Breast Cancer and Their Clinical Implications: A Gene-Expression-Based Retrospective Study. PLoS medicine. 13-1002194.
- 107.Nirmal A J, Regan T, Shih B B, Hume D A, Sims A H et al. (2018) Immune Cell Gene Signatures for Profiling the Microenvironment of Solid Tumors. Cancer immunology research. 6, 1388-1400.
- 108.Ichinohe T, Miyama T, Kawase T, Honjo Y, Kitaura K et al. (2018) Next-Generation Immune Repertoire Sequencing as a Clue to Elucidate the Landscape of Immune Modulation by Host-Gut Microbiome Interactions. Frontiers in immunology. 9-668.
- 109.Chifman J, Pullikuth A, Chou J W, Bedognetti D, Miller L D. (2016) Conservation of immune gene signatures in solid tumors and prognostic implications. BMC cancer. 16-911.
- 110.Schelker M, Feau S, Du J, Ranu N, Klipp E et al. (2017) Estimation of immune cell content in tumour tissue using single-cell RNA-seq data.NatureCommunications. 8:.
- 111.Zou J, Wang E eTumorType. (2017) An Algorithm of Discriminating Cancer Types for Circulating Tumor Cells or Cell-free DNAs in Blood. Genomics, proteomics & bioinformatics. 15, 130-140.
- 112.Chen S H, Kuo W Y, Su S Y, Chung W C, Ho J M et al. (2018) A gene profiling deconvolution approach to estimating immune cell composition from complex tissues. BMC bioinformatics. 19, 154.
- 113.Chen Z, Quan L, Huang A, Zhao Q, Yuan Y et al. (2018) seq-ImmuCC: Cell-Centric View of Tissue Transcriptome Measuring Cellular Compositions of Immune Microenvironment From Mouse RNA-Seq Data. Frontiers in immunology. 9, 1286.
Cited by (1)
- 1.Zhang Xi, Moore Camille M., Harmacek Laura D., Domenico Joanne, Rangaraj Vittobai Rashika, et al, 2022, CFTR-mediated monocyte/macrophage dysfunction revealed by cystic fibrosis proband-parent comparisons, JCI Insight, 7(6), 10.1172/jci.insight.152186
